Smntx Tutorials
- Using the Smntx GUI: This guide includes an overview of the Smntx GUI and demonstrates how to start an NLP analysis, view the results, mine multiple documents and execute different queries.
- Accessing Smntx through Taverna: This document shows how scientific medical NLP workflows can be created using the Smntx REST interface and Taverna.
Accessing Smntx through Taverna
The standardised nature of the Smntx REST API makes it easily consumable by a wide variety of tools, for example language independent applications and workflows. This tutorial describes the process of creating a scentific Taverna based workflow to automate medical text analysis using MetaMap.
In order to follow this tutorial you will need:
- Access to the MetaMap REST API, the URL of which is referred to as <METAMAP_URL>
- A preconfigured username to access the MetaMap REST service
- Taverna 2.2 (http://www.taverna.org.uk/)
- Taverna REST Activity Plugin
The aim of this tutorial is to construct a workflow that automatically analyzes medical text for a particular semantic type (e.g diseases). Given this input text and a specific semantic type, the workflow will return the UMLS concept names that match the semantic type and also the sentences in which the semantic type occurs.
- Start a new analysis:
- Create a beanshell to construct an JSON object:
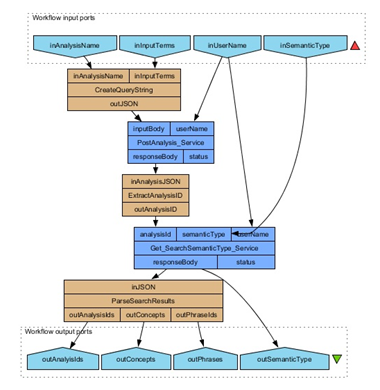
In order to start analysis you will first need to create a beanshell processor to construct the JSON Analysis object. Create a beanshell with two input ports (inAnalysisName, inInputTerms) and one output (outJSON). Enter a single line of code in the beanshell to form the JSON object. Note: interactive is set to true so that the next stage of the workflow will wait for the analysis to complete.
String outJSON = "{\"applicationName\":\"" + inAnalysisName + "\", \"inputTerms\":\"" + inInputTerms + "\", \"interactive\":\"true\"}"; - Create a REST service processor to start the analysis:
Create a REST service using the REST Activity plugin. Name the service POST_CreateAnalysis_Service. Select POST for the HTTP Method, select application/json for both the accept and content-type header, select send data as binary, and use the following URL for the URL template.
String outJSON = "{\"applicationName\":\"" + inAnalysisName + "\", \"inputTerms\":\"" + inInputTerms + "\", \"interactive\":\"true\"}";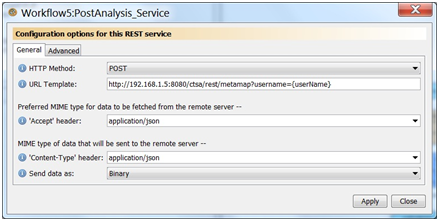
You should now have a new processor with 2 inputs (inputBody, username) and 2 outputs.
- Add input ports and connect up processors:
Add three input ports to the workflow (inAnalysisName, inInputTerms, and inUserName) so users can change arguments when running the workflow. Connect the first two inputs to the beanshell, connect the output of the beanshell (outJSON) to the inputBody of the REST service and inUserName to the userName input of the REST service.
- Test the workflow by starting a new analysis :
To test the workflow, add a workflow output port (outInstance), link the responseBody output of POST_CreateAnalysis_Service to the workflow output port (outInstance) and execute the workflow with the following parameters:
inAnalysisName = “first test”
inInputTerms = “The patient might have cancer.”
inUserName = “<USERNAME>'”
This should result in the JSON analysis object being displayed for the output port, the output should look similar to:
{"id":"751", "applicationName":"firsttest", "instancePath":"C:\\UC\\CTSASupplement\\Temp\\Kyle\\751", "inputTerms":"The patient might have cancer.", "interactive":"true", "date":"1290549012051", "status": "Complete", "markedupResultId":"801"}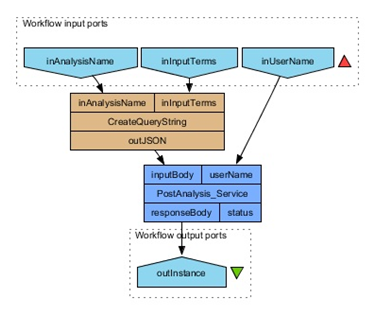
- Create a beanshell to construct an JSON object:
- Filter results based on semantic type:
There are two ways to search for semantic type using the MetaMap REST API, first the analysis can be run with a filter applied such that only the required semantic types are returned, or secondly a user based search can be run after the analysis is complete. For this tutorial we use the second approach as this can be customized and run independently with different search criteria without re-executing the analysis.
- Add semantic type input port:
Add an additional workflow input port (inSemanticType) so that users can specify the semantic type they want to find.
- Create a REST service processor to search the results for a given semantic type:
Add the REST service to the workflow. Name the service GET_SearchSemanticType_Service. Select GET for the HTTP Method, select application/json for the accept header, and use the following URL for the URL template.
http://192.168.1.5:8080/ctsa/rest/search? username={userName}&semantictype_exact={semanticType}&instanceid={analysisId}&mapping=true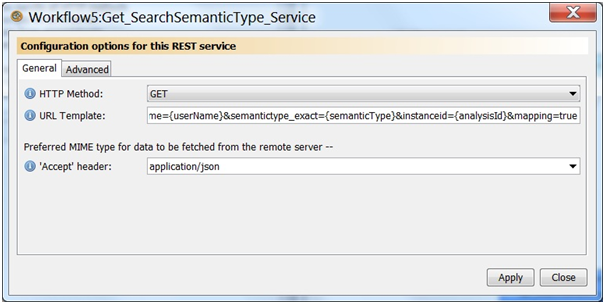
This should create a new service with three input ports (analysisId, semanticType, and username).
- Add a beanshell to get the analysis ID from the completed analysis:
Create a beanshell processor to extract the instance ID from the analysis JSON object. Create a single input port (inAnalysisJSON) and a single output port (outAnalysisID). Enter the following code to get the id from the input JSON using string based operations. Note this code contains no error checking.
String outAnalysisID = "";
if (instanceJSON != null){
String [] values = inAnalysisJSON.split(",");
String [] id = values[0].split(":");
outAnalysisID = id [1];
}
outAnalysisID = outAnalysisID.replaceAll("\"", "");
- Link the semantic type REST service with the input ports and beanshell:
Connect the output of the beanshell (outAnalysisID) to the input port (analyisId) of the search REST service. Connect the other two input ports (semanticType and username) to the appropriate workflow input ports.
- Add an output port and test the workflow:
You can now run the workflow and retrieve raw search results for the specified query. For example to find all mappings that are classified as neoplastic processes (neop), enter the following input port values and execute the workflow.
inAnalysisName = “second test”
inInputTerms = “The patient might have cancer.”
inUserName = “<USERNAME>”
inSemanticType = “neop”
This should return the search output with three results that match the query. The first result should look similar to:
...[{"username":"Kyle", "userid":"1", "analysisname":"secondtest", "instanceid":"756", "patientgroupid":"1", "patientgroupname": "Ungrouped", ”phraseid":"2", "phrase":"cancer.", "mapping":true, "conceptname":"Cancer", "preferredname":"Malignant Neoplasms", "mmscore":-1000, "id":"4798ac5a-094d-410c-8a00", "semantictype":["neop"], "source":["MTH","PSY",...]}...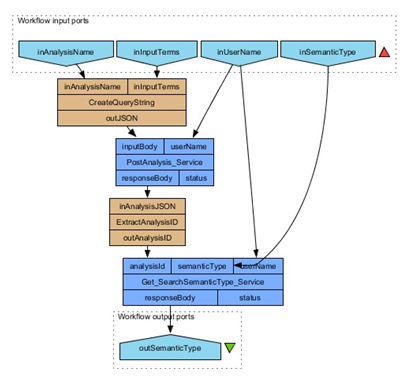
- Add semantic type input port:
The search results returned are somewhat raw, in order to be able to take what is needed from these results a beanshell processor can be used to parse the results. For example assume you are looking for a list of UMLS concepts and also unique phrase ids (to retrieve sentence context). To do this create a beanshell called ParseSearchResults with one input port (inJSON) and 3 output ports (outConcepts, outPhraseIds, outAnalysisIds) ensure the output ports are of depth 1 (as there are multiple values for each one in the search results). Note: you will also need to ensure the Jackson JSON jar is accessible to Taverna. Enter the following code in the beanshell.
import org.codehaus.jackson.map.ObjectMapper;
ObjectMapper mapper = new ObjectMapper();
List outConcepts = new ArrayList();
List outAnalysisIds = new ArrayList();
List outPhraseIds = new ArrayList();
try{
JsonNode rootNode = mapper.readValue(inJSON, JsonNode.class);
JsonNode results = rootNode.findValue("docs");
Iterator it = results.getElements();
JsonNode doc;
while (it.hasNext()){
doc = (JsonNode)it.next();
outConcepts.add(doc.get("conceptname").getTextValue());
outPhraseIds.add(doc.get("phraseid").getTextValue());
outAnalysisIds.add(doc.get("instanceid").getTextValue());
}
} catch (Exception e){
System.out.println("Exception " + e);
}
Connect the output of the Get_SearchSemanticType_Service processor to the input of the new beanshell. Create two new output ports to check the parsing of the search results (outConcepts, outPhrases). Re-execute the workflow. You should see a list of output for both the outConcept and outPhrases output ports. Because of the trivial nature of the input terms the three results are all the same (cancer).
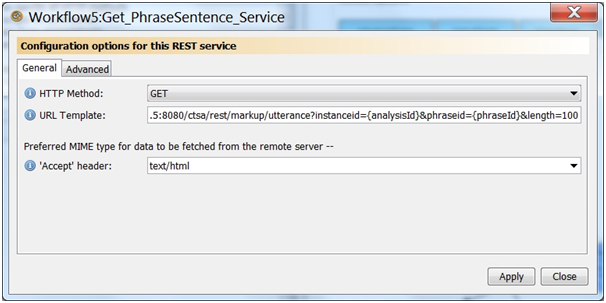
Add a new REST service processor to get the utterance, name the service Get_PhraseSentence_Service. Specify the following properties for the new REST service HTTP Method: GET, Accept header: text/html, and URL Template:
Link up the output ports (outAnalysisI, outPhraseIds) from the parseSearchResults beanshell to the input ports of the new Get_PhraseSentence_Service (analysisId, phraseId). Link the output port (responseBody) to a new output Port called outSentences.
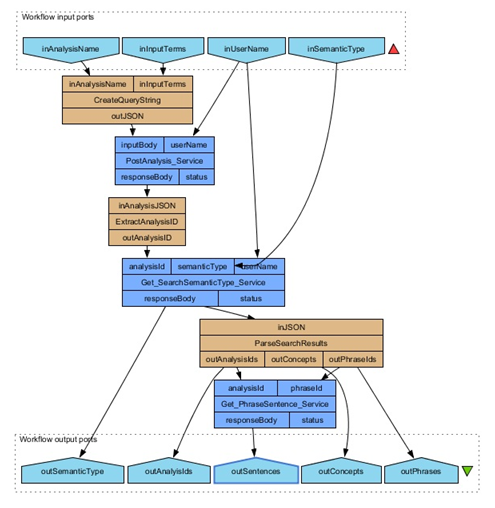
You have now completed the workflow. Run the workflow with the following parameters.
- inAnalysisName = “second test”
- inInputTerms = “The patient might have cancer.”
- inUserName = “<USERNAME>”
- inSemanticType = “neop”
You should then be able to view the results.
- outConcepts: is a list of all UMLS concepts that match the search criteria of semantic type ‘neop’ in the input terms
- outSemanticType: is the raw JSON search results of the query.
- outSentences: is a list of all the sentences in which the matching phrases occurred. Phrases are wrapped in an HTML span tag to be displayed in a browser.